Lec 11 Backpropagation with Spatiotemporal Adjustment for Training SNNs Based on BrainCog
Spiking Neural Network (SNN), is closer to the information processing structure and mechanism of human brain, uses spike sequences to transmit information has richer spatio-temporal information processing capability than artificial neural networks. However, how to train SNNs more efficiently in order to obtain more efficient learning and information processing capabilities has been a concern of researchers. The introduction of Surrogate Gradient helps the training of SNNs, but it has also brought problems. Most researchers tend to simply consider SNNs as a substitute for Recurrent Neural Network (RNN) and train directly using BPTT (Backpropagation Through Time) algorithms Directly applying the backpropagation algorithm to spiking neural network training can neither exploit the advantages of backpropagation nor lose the significance of using spiking neurons for intelligent information processing.
In order to make the training of spiking neural network more efficient, we propose a more bio-rational spatio-temporal back-propagation algorithm for training deep spiking neural networks. By rethinking and reshaping the relationship between neuronal membrane potentials and spikes in the spatial dimension, the algorithm achieves a regulation of gradient back-propagation in the spatial dimension, avoiding the unnecessary influence of non-spiking neurons on weight. The temporal dependence problem of traditional spiking neurons within a single spike cycle is overcome by biologically adjustment of the gradient in the temporal dimension, which helps the error to be transmitted across multiple spikes and enhances the temporal dependence of SNNs.
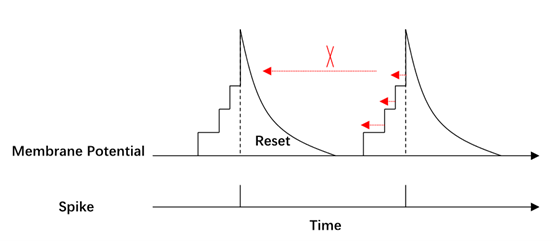
The membrane potential of spiking neurons changes as a process of information accumulation. After the neurons have accumulated enough information, they will send the information to the post-synaptic neurons in the form of spikes. As a result, the binary spikes can be regarded as a normalization of the information contained in the membrane potential. For the BP process, it is more reasonable to only calculate the gradient of the neuron at the moment of spiking to the membrane potential. We propose the BPSA to improve the training of SNNs. When the membrane potential does not reach the threshold, we will clip the gradient of the spikes to the membrane potential to avoid the problems of repeated updates at an earlier time, as in Figure 4. When the membrane potential reaches the threshold, we normalize the membrane potential and spread the information in spikes.
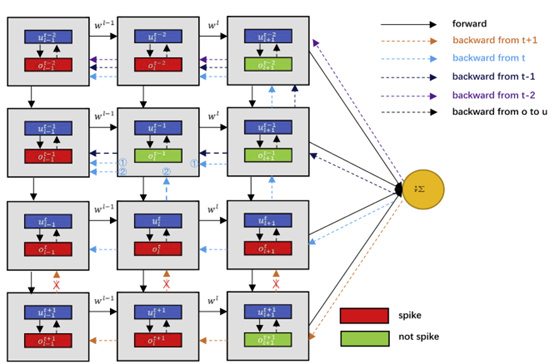
In biological neurons, the spike that the neuron fires will affect the subsequent spikes of the neuron. When directly using the BP algorithm to optimize the parameter, the gradient of the loss function to the neuron output will only be propagated from the time the neuron was last excited to the present and will not cross the spikes as shown in Figure 5. So, the influence between spikes will not be considered in the temporal dimension. Then, we propose a BPTA cross the spikes. Considering that the temporal dependence disappears during the BP process, we add the residual connection between spikes during the backward, as shown in Figure 6.
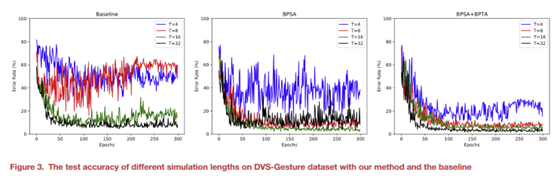
The latency is one of the main problems that restricts the development of SNNs. The spiking neurons need to accumulate membrane potential, and once they reach the threshold, they fire spikes and transmit information. Therefore, SNNs often require a long simulation time to achieve higher performance. Here, we study the influence of different simulation lengths on the network performance. As shown in the Figure 3, when our adjustments are not introduced, when the simulation time is reduced, the test curve of the network is not very smooth. As can be seen in Table 6, with the introduction of the two adjustments, our training method still achieves high accuracy while reducing the simulation time.
This biologically plausible spatio-temporal modulation is integrated in BrainCog and can be easily applied to neuromorphic data classification tasks. You can refer to the previous tutorials for details on how to use it. Thank you all!
class HTDGLIFNode(IFNode):
def __init__(self, threshold=.5, tau=2., *args, **kwargs):
super().__init__(threshold, *args, **kwargs)
self.warm_up = False
def calc_spike(self):
spike = self.mem.clone()
spike[(spike < self.get_thres())] = 0.
# self.spike = spike / (self.mem.detach().clone() + 1e-12)
self.spike = spike - spike.detach() + \
torch.where(spike.detach() > self.get_thres(), torch.ones_like(spike), torch.zeros_like(spike))
self.spike = spike
self.mem = torch.where(self.mem >= self.get_thres(), torch.zeros_like(self.mem), self.mem)
self.mem = (self.mem + 0.2 * self.spike - 0.2 * self.spike.detach()) * self.dt
def forward(self, inputs):
if self.warm_up:
return F.relu(inputs)
else:
return super(IFNode, self).forward(F.relu(inputs))
Welcome to refer and cite our papers.
@misc{https://doi.org/10.48550/arxiv.2207.08533,
doi = {10.48550/ARXIV.2207.08533},
url = {https://arxiv.org/abs/2207.08533},
author = {Zeng, Yi and Zhao, Dongcheng and Zhao, Feifei and Shen, Guobin and Dong, Yiting and Lu, Enmeng and Zhang, Qian and Sun, Yinqian and Liang, Qian and Zhao, Yuxuan and Zhao, Zhuoya and Fang, Hongjian and Wang, Yuwei and Li, Yang and Liu, Xin and Du, Chengcheng and Kong, Qingqun and Ruan, Zizhe and Bi, Weida},
title = {BrainCog: A Spiking Neural Network based Brain-inspired Cognitive Intelligence Engine for Brain-inspired AI and Brain Simulation},
publisher = {arXiv},
year = {2022}
}
@article{shen2022backpropagation,
title = {Backpropagation with biologically plausible spatiotemporal adjustment for training deep spiking neural networks},
author = {Shen, Guobin and Zhao, Dongcheng and Zeng, Yi},
journal = {Patterns},
pages = {100522},
year = {2022},
publisher = {Elsevier}
}